PDF) A fast and interpretable deep learning approach for accurate electrostatics-driven pKa predictions in proteins


PDF) A fast and interpretable deep learning approach for accurate electrostatics-driven pKa predictions in proteins

A Fast and Interpretable Deep Learning Approach for Accurate Electrostatics-Driven pKa Predictions in Proteins

A Review on Predicting Drug Target Interactions Based on Machine Learning

Network biology and artificial intelligence drive the understanding of the multidrug resistance phenotype in cancer - ScienceDirect
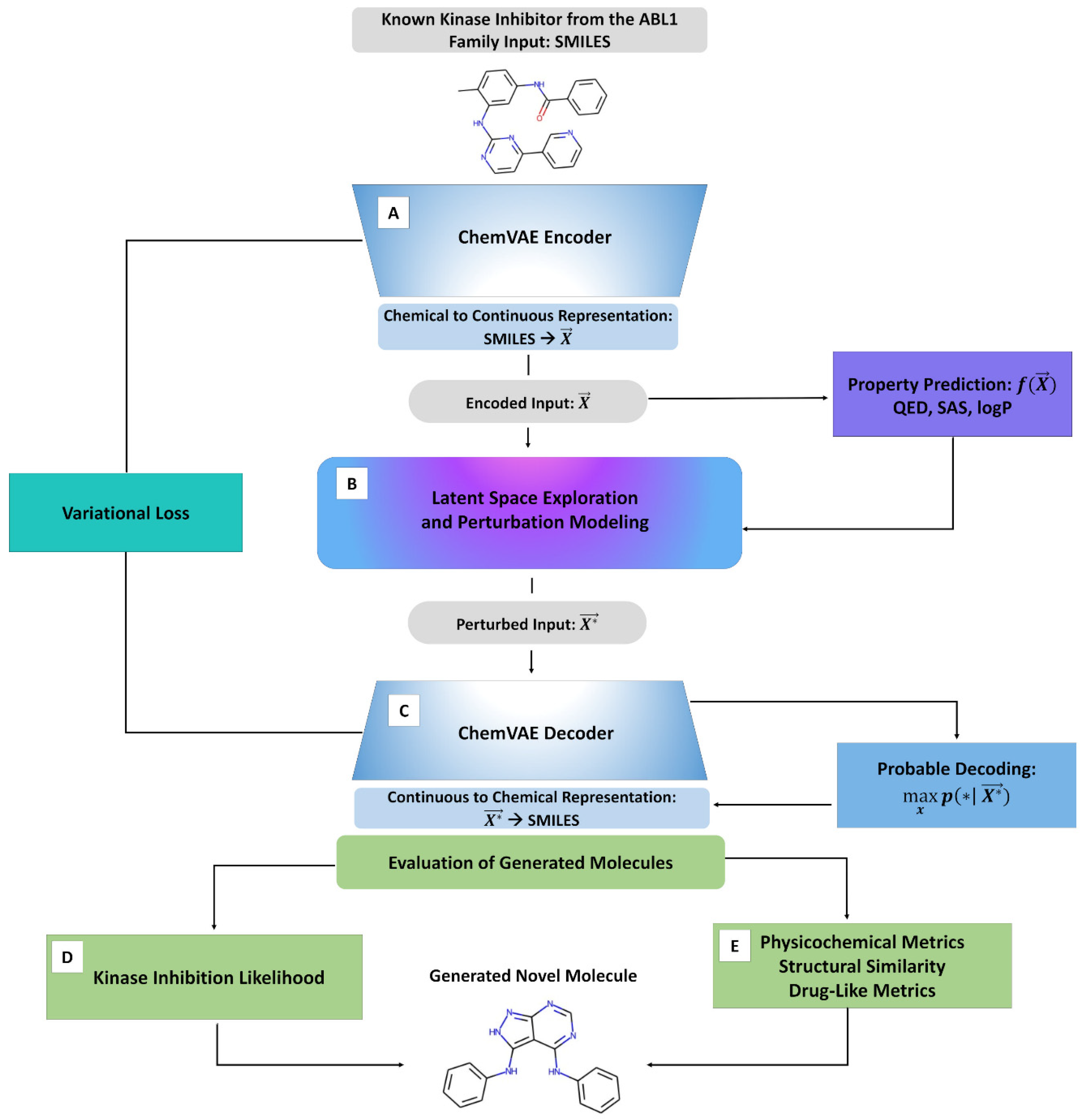
IJMS, Free Full-Text

Prediction Machines: Applied Machine Learning for Therapeutic Protein Design and Development - Journal of Pharmaceutical Sciences

A Fast and Interpretable Deep Learning Approach for Accurate Electrostatics-Driven pKa Predictions in Proteins
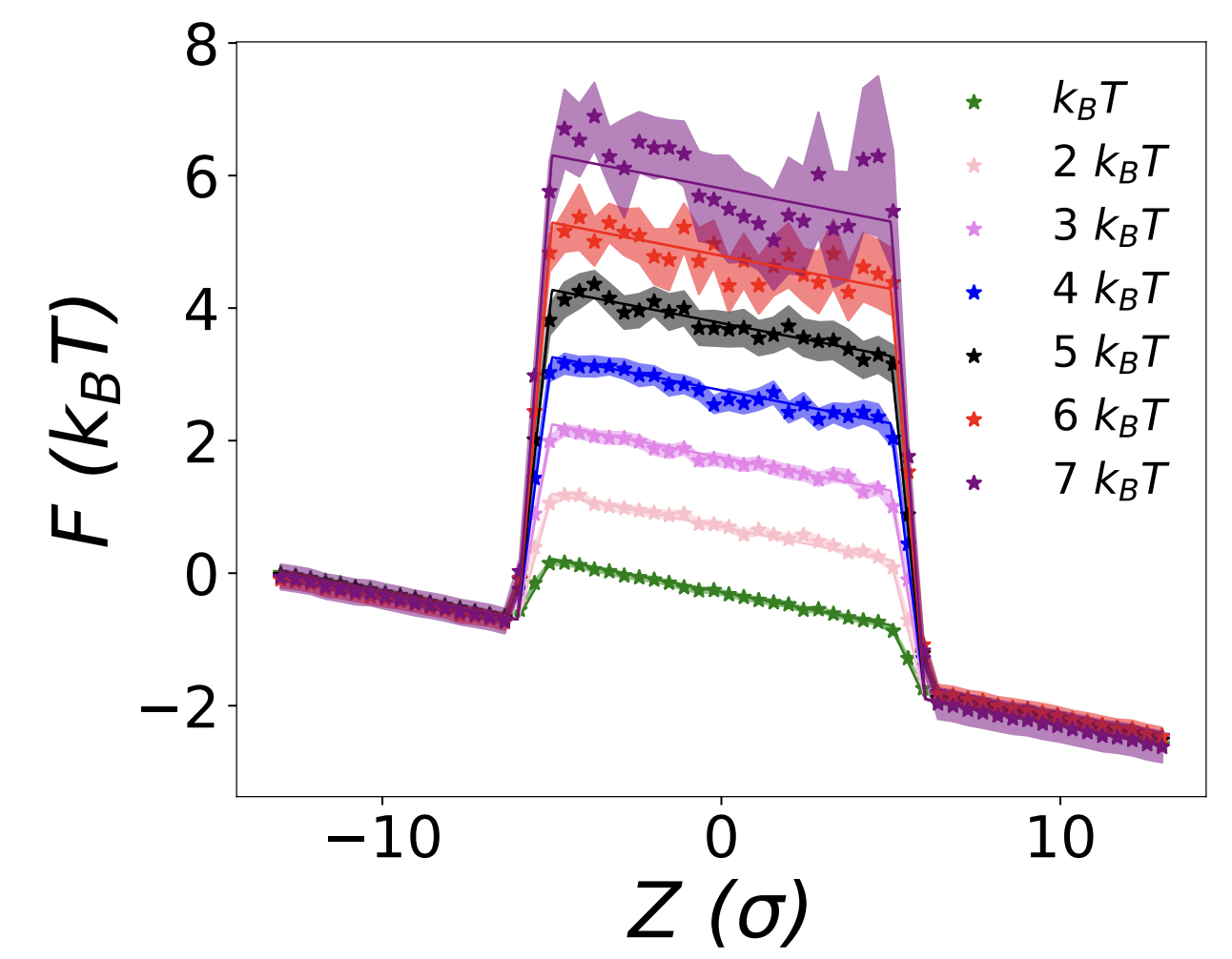
Midwest Integrated Center for Computational Materials - Publications

Machine learning methods for pKa prediction of small molecules: Advances and challenges - ScienceDirect

Prediction Machines: Applied Machine Learning for Therapeutic Protein Design and Development - Journal of Pharmaceutical Sciences

Biomolecules, Free Full-Text

PDF] PKAD: a database of experimentally measured pKa values of ionizable groups in proteins









